A transformer-based molecular docking paradigm for large-scale virtual screening
In this study, a novel transformer-based architecture named Dockformer is proposed to overcome the above-mentioned issues of current DL-based docking methods.
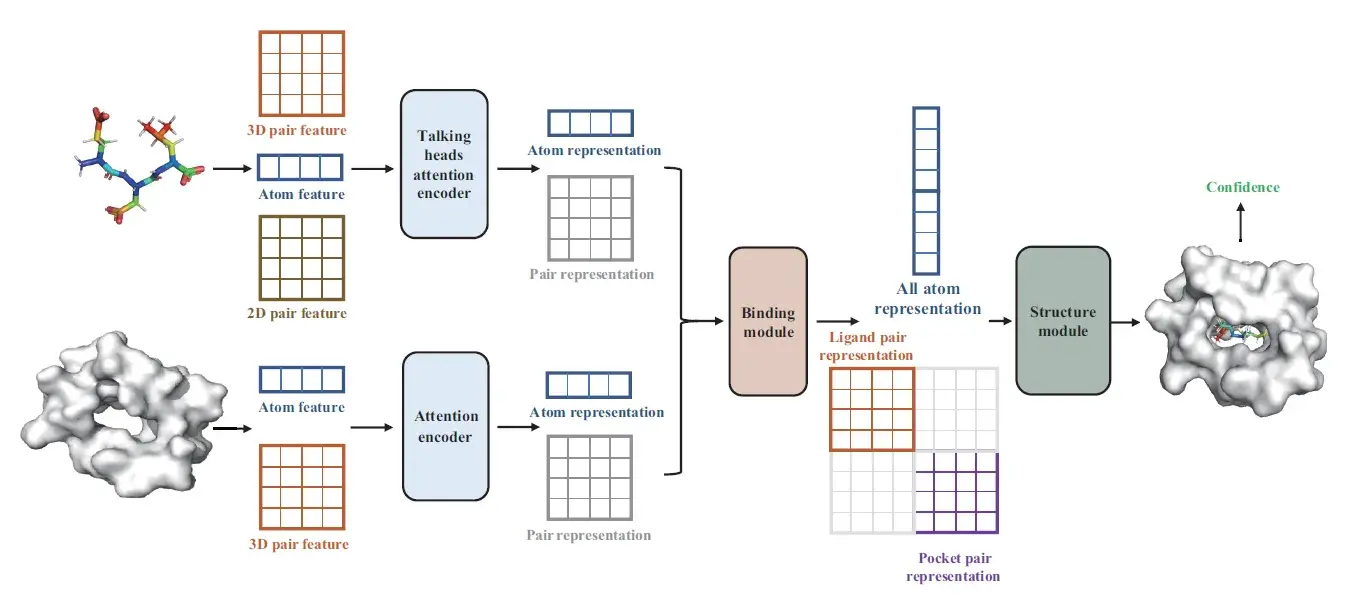
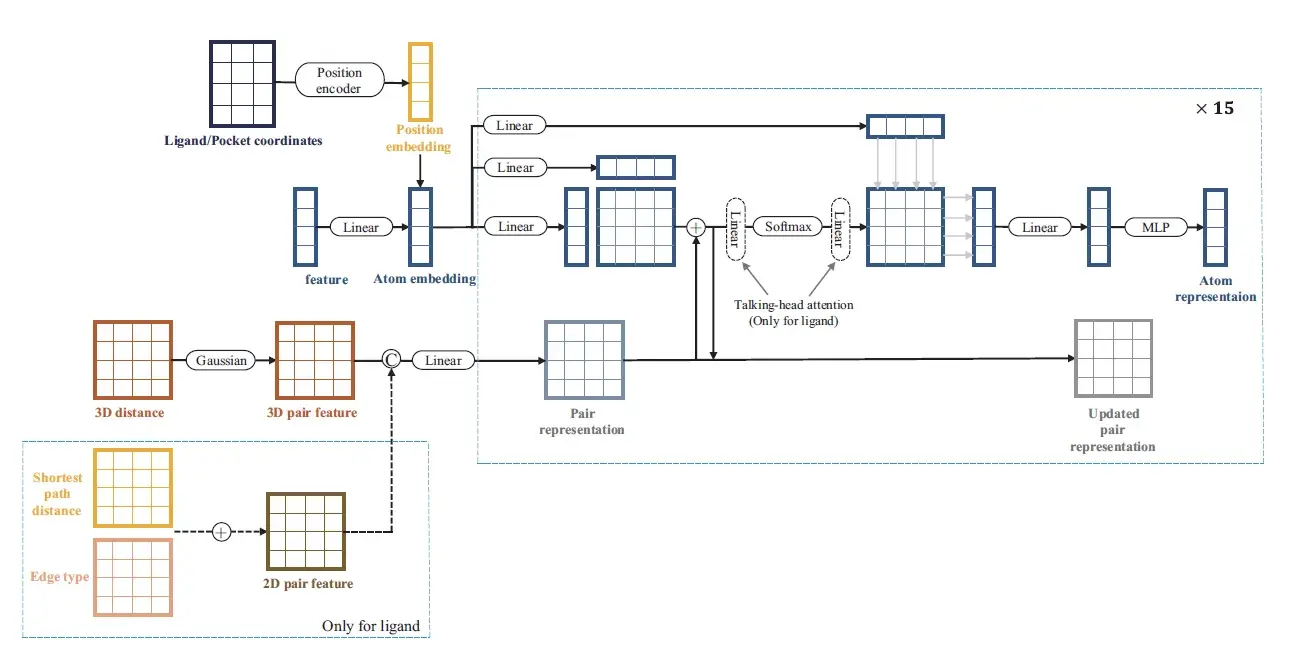
Specifically, Dockformer uses two separate transformer encoders to leverage multimodal information to generate latent embeddings of proteins and ligands and can thus effectively capture molecular geometric details, including 2D graph topology and 3D structural knowledge. To strengthen the transformer’s spatial sensitivity, we augment atom representations with 3D point positional encodings, furnishing rich geometric context for subsequent structure prediction.
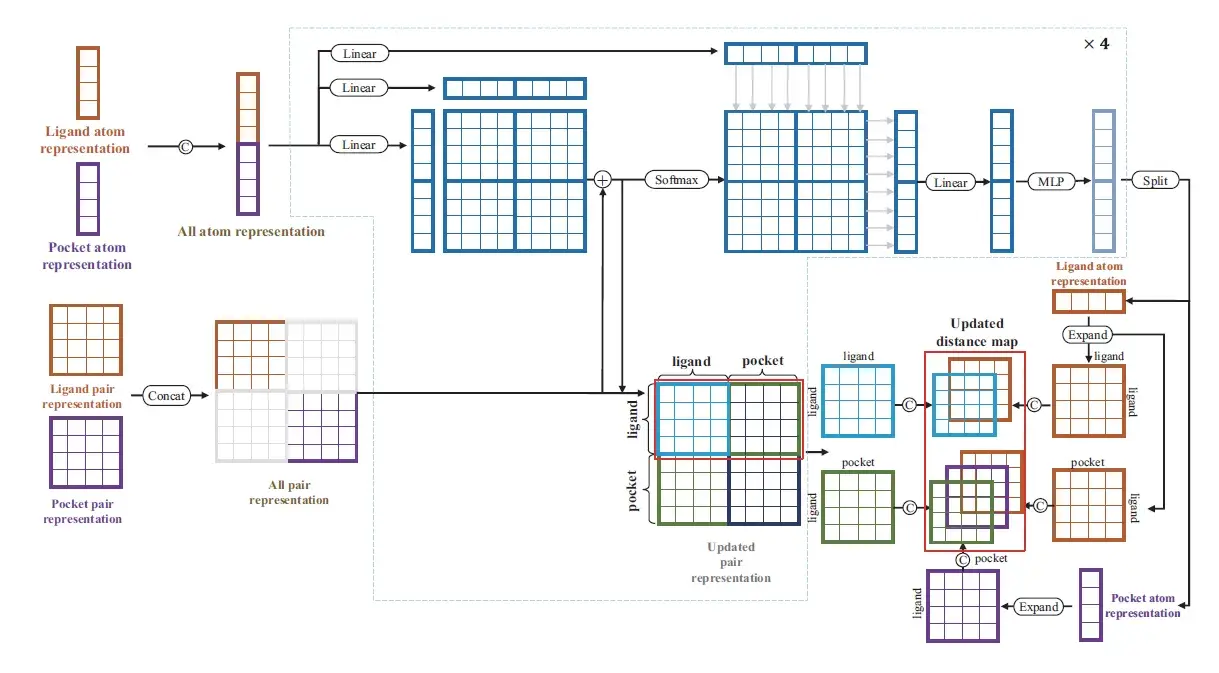
Next, a binding module is then employed to detect intermolecular relationships effectively on the basis of learned latent embeddings.
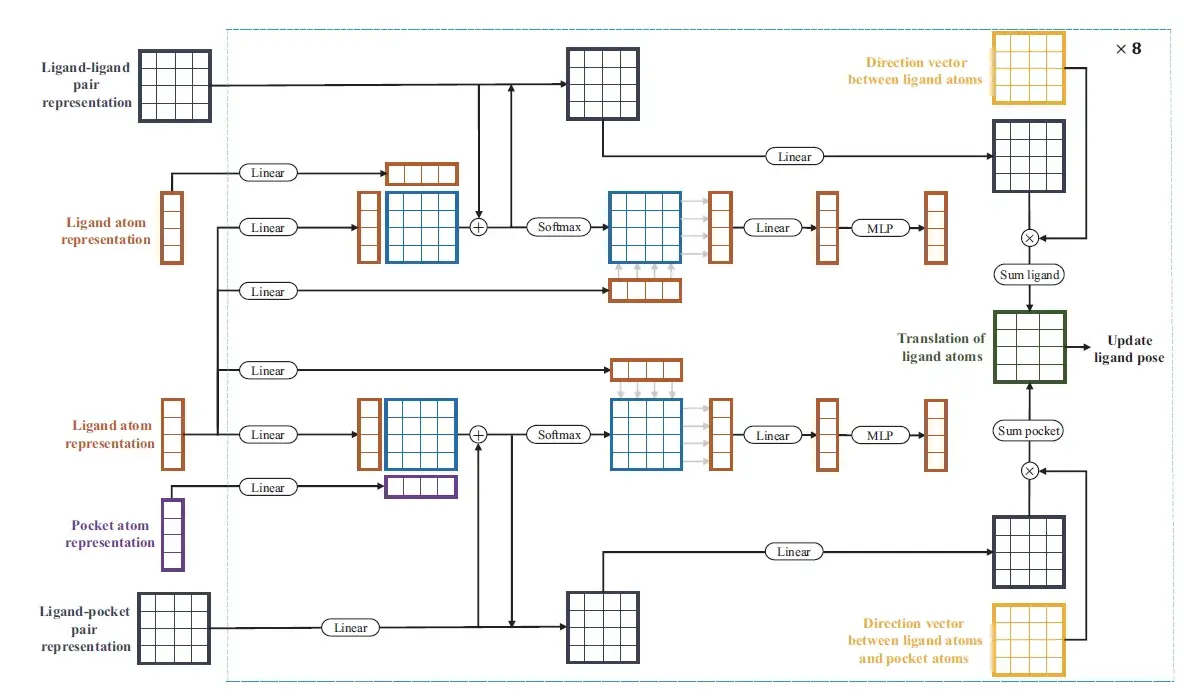
Finally, in the structure module, the established relationships are utilized to generate the complex conformations directly, and the coordinates of the ligand atoms are calculated in an end-to-end manner. In addition, the corresponding confidence measures of each generated conformation are utilized to distinguish binding strengths instead of traditional scoring functions.
In summary, distinct from conventional DL-based and optimization-based docking methods, the multimodal information fusion equips Dockformer with superior docking accuracy, and the end-to-end architecture enables it to simultaneously speed up the conformation generation process by orders of magnitude. Thus, this method can meet the rapid throughput requirements of LSVS tasks. Dockformer, as a robust and reliable proteinligand docking approach, may significantly reduce the development cycle and cost of drug design.
Papers
- Zhangfan Yang, Junkai Ji, Shan He, Jianqiang Li, Tiantian He, Ruibin Bai, Zexuan Zhu, Yew Soon Ong. “Dockformer: A transformer-based molecular docking paradigm for large-scale virtual screening.” arXiv preprint arXiv:2411.06740 (2024). View Paper